Find Sample Size for RMST Model with Dependent Censoring (Analytic)
Source:R/dependent_censoring_analytical.R
DC.ss.analytical.RdCalculates the required sample size for a target power for an RMST model with dependent censoring.
Usage
DC.ss.analytical(
pilot_data,
time_var,
status_var,
arm_var,
dep_cens_status_var,
target_power,
linear_terms = NULL,
L,
alpha = 0.05,
n_start = 50,
n_step = 25,
max_n_per_arm = 2000
)Arguments
- pilot_data
A
data.framecontaining pilot study data.- time_var
A character string for the time-to-event variable.
- status_var
A character string for the primary event status (1=event, 0=otherwise).
- arm_var
A character string for the treatment arm variable (1=treatment, 0=control).
- dep_cens_status_var
A character string for the dependent censoring status (1=dependent event, 0=otherwise).
- target_power
A single numeric value for the desired power.
- linear_terms
An optional character vector of other covariate names.
- L
The numeric value for the RMST truncation time.
- alpha
The significance level (Type I error rate).
- n_start
The starting sample size per arm for the search.
- n_step
The increment in sample size at each step of the search.
- max_n_per_arm
The maximum sample size per arm to search up to.
Value
A list containing:
- results_data
A
data.framewith the target power and required sample size.- results_plot
A
ggplotobject visualizing the search path.- results_summary
A
data.framesummarizing the estimated treatment effect.
Details
This function performs an iterative search for the sample size needed to
achieve a specified target_power. It uses the same underlying theory as
DC.power.analytical. It performs a one-time estimation of the treatment
effect and its asymptotic variance from the pilot data, then uses these
parameters in an analytic formula to efficiently search for the required sample size.
Examples
# Generate sample pilot data with a clear treatment effect
set.seed(456)
n_pilot <- 200
pilot_df_ss <- data.frame(
time = rexp(n_pilot, rate = 0.2),
arm = rep(0:1, each = n_pilot / 2),
age = rnorm(n_pilot, mean = 55, sd = 8)
)
# Introduce a treatment effect
pilot_df_ss$time[pilot_df_ss$arm == 1] <- pilot_df_ss$time[pilot_df_ss$arm == 1] * 1.5
# Create competing event indicators
event_type <- sample(0:2, n_pilot, replace = TRUE, prob = c(0.6, 0.2, 0.2))
pilot_df_ss$status <- ifelse(event_type == 0, 1, 0)
pilot_df_ss$dep_cens_status <- ifelse(event_type == 1, 1, 0)
pilot_df_ss$time[event_type != 0] <- pilot_df_ss$time[event_type != 0] * 0.7
# Run the sample size search
dc_ss_results <- DC.ss.analytical(
pilot_data = pilot_df_ss,
time_var = "time",
status_var = "status",
arm_var = "arm",
dep_cens_status_var = "dep_cens_status",
target_power = 0.80,
linear_terms = "age",
L = 15,
alpha = 0.05,
n_start = 100,
n_step = 50
)
#> --- Estimating parameters from pilot data for analytic calculation... ---
#> Model: Y_rmst ~ arm + age
#> --- Searching for Sample Size (Method: Analytic) ---
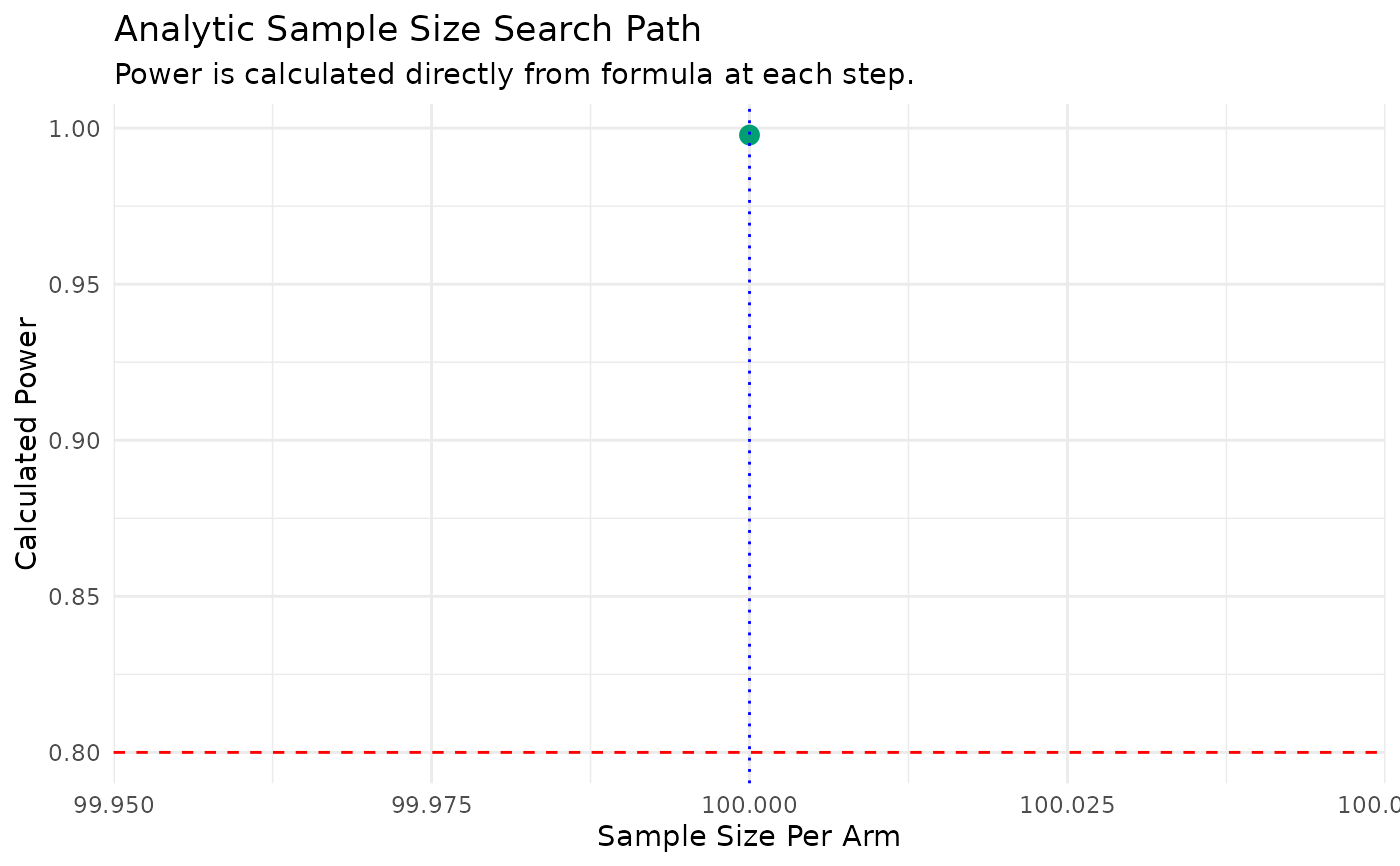
#> N = 100/arm, Calculated Power = 0.998
#>
#> --- Calculation Summary ---
#>
#>
#> Table: Required Sample Size
#>
#> | Target_Power| Required_N_per_Arm|
#> |------------:|------------------:|
#> | 0.8| 100|
print(dc_ss_results$results_data)
#> Target_Power Required_N_per_Arm
#> 1 0.8 100
print(dc_ss_results$results_plot)
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?